Goals
- Identify spatial clusters with significantly low immunization rates
- Develop an agent-based model for the spread of measles that incorporates detailed immunization data, and is calibrated using a novel source of incidence data
- Develop methods to find and characterize critical spatial clusters, with respect to different metrics, which capture both epidemic and economic burden, and order underimmunized clusters based on their criticality
- Use the methodology to evaluate interventions in terms of their effect on criticality
One fundamental task in network analysis is detecting “hotspots” or “anomalies” in the network; that is, detecting subgraphs where there is significantly more activity than one would expect given historical data or some baseline process. Scan statistics is one popular approach used for anomalous subgraph detection. This methodology involves maximizing a score function over all connected subgraphs, which is a challenging computational problem. A number of heuristics have been proposed for these problems, but they do not provide any quality guarantees. Here, we provide a framework for designing algorithms for optimizing a large class of scan statistics for networks, subject to connectivity constraints. Our algorithms run in time that scales linearly on the size of the graph and depends on a parameter we call the “effective solution size,” while providing rigorous approximation guarantees. In contrast, most prior methods have super-linear running times in terms of graph size. Extensive empirical evidence demonstrates the effectiveness and efficiency of our proposed algorithms in comparison with state-of-the-art methods. Our approach improves on the performance relative to all prior methods, giving up to over 25% increase in the score. Further, our algorithms scale to networks with up to a million nodes, which is 1--2 orders of magnitude larger than all prior applications.
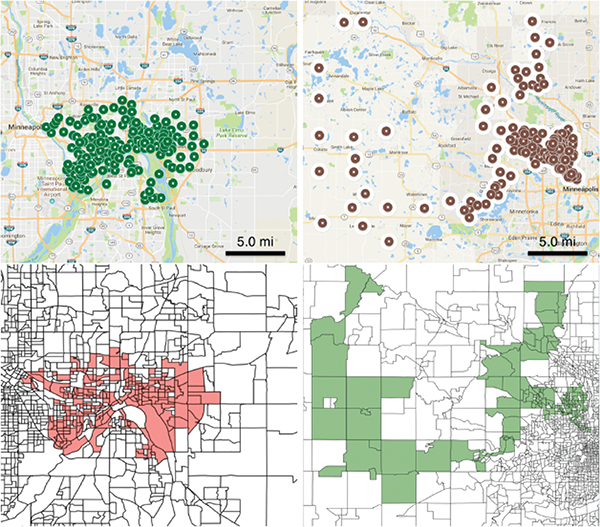
Emergence of under-immunized clusters is a growing concern for public health agencies because they can act as reservoirs of infection and increase the risk of infection into the wider population. We use realistic population models for Minnesota and Washington state, and combine this with school level immunization data to estimate vaccine coverage at the level of census block groups. A scan statistic method defined on networks is used for finding significant clusters of under-immunized block groups, without any restrictions on shape. Further we provide the demographic characteristics of these clusters. We find significant under-vaccinated clusters in MN and WA. These are very irregular in shape, in contrast to the circular disks reported in prior work, which rely on the SatScan approach. Some of the clusters found by our method are not contained in those computed using SatScan, a state-of-the-art software tool used in similar studies in other states. Higher resolution clusters computed using our network based approach and population models provide new insights on the structure and characteristics of such clusters and enable targeted interventions (Cadena et al. BMC Medical Informatics and Decision-Making).
Figures
Other Team Members
Travis Porco | Professor of Global Health | University of California, San Francisco
Nicola Klein | Senior Research Scientist | Kaiser Permanente