Understanding Large-Scale Networks
Researchers studied the complexity arising from large-scale networks. One particular example is long non-coding RNAs (lncRNAs, typically 1-20 kB) viewed as contact graphs. Recently, lncRNAs have been found to be pervasively transcribed in the human and other mammalian genomes, and are increasingly associated with networks of epigenetic and post-transcriptional control in healthy and diseased biological systems. Understanding the secondary structure of lncRNAs is the key to unveiling its diverse functional roles in gene regulation. In particular, long-range intramolecular interactions in these large molecules are poorly understood and present a significant challenge to predicting secondary structures for lncRNAs.
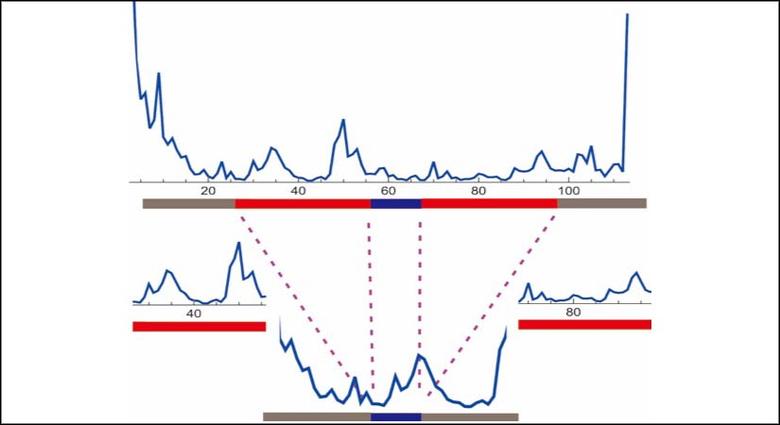
We investigate the Boltzmann ensemble of secondary structures generated by statistical sampling, and identify the structural features by extracting the information of such an ensemble. We utilize an integrated experiment and computation approach to understand the secondary structure of lncRNAs. Our research facilitates the discovery of the regulation role of lncRNAs and further implications for molecular markers of diseases such as cancer.
Research Highlights
- We analyzed the distributions of various structural elements in lncRNAs, such as stacks, hairpin- , interior- and multi-loops, and pseudoknots.
- We analyzed the length spectrum of base-pairings in large RNA secondary structures. The long-range base pairings have length on the order of the sequence length.
- We developed a computational approach to identify the key features of base-pairing interactions, by extracting information from large-scale statistical sampling of secondary structures.
Ongoing Research
- We are currently working on developing an information-theoretic framework that incorporates key structural features and experimental probing data in a hierarchical fashion to determine the secondary structure of lncRNAs.
- We further studied the relationship between the networks of base-pair interactions in the ensemble and the dynamics generated by sequential update of information from experimental probing data.
Tools
-
 This is a software package for identifying the target secondary structure from an RNA structure ensemble. We present a demo of the ensemble tree.
This is a software package for identifying the target secondary structure from an RNA structure ensemble. We present a demo of the ensemble tree.