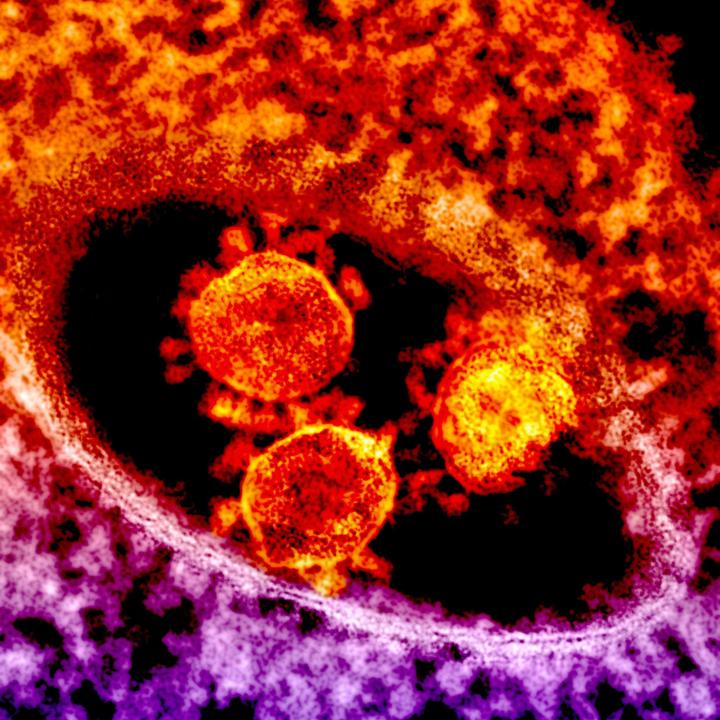
The Bacterial and Viral Bioinformatics Resource Center (BV-BRC) is an information system designed to support the biomedical research community’s work on bacterial and viral infectious diseases by integrating vital pathogen information with rich data and analysis tools. BV-BRC combines the data, technology, and extensive user communities from two long-running centers: PATRIC, the bacterial system, and IRD/ViPR, the viral systems. In addition to nearly 1 million bacterial genomes and 9 million viral genomes, BV-BRC provides host data on protein structure and function, clinical studies, drug targets and resistance, epidemiology, and other features. As well, it offers open-source tools for data analysis and genomic annotation.
BV-BRC and its predecessor systems have over 40,000 registered users, tens of thousands of unique visitors per month, over 18,000 citations, and over 100 publications.
Infectious pathogens in bacteria, viruses, fungi, and parasites naturally evolve over time. As they do, they can develop antimicrobial resistance (AMR) to available medicines. When this happens, antibiotics and other antimicrobial medicines lose efficacy and these infections become more difficult to treat, increasing the likelihood of disease spread, severity, and death.
The Bacterial and Viral Bioinformatics Resource Center (BV-BRC), a beta website that launched in February, “is one-stop shopping for your bacterial or viral genomic research,” said Ron Kenyon, a senior scientist in the Network Systems Science and Advanced Computing division of UVA’s Biocomplexity Institute.